We have developed a model system for the study of akinetes in this organism using a metabolic mutant lacking glucose-6-phosphate dehydrogenase (encoded by the zwf gene). The cells of this mutant synchronously turn into akinetes after several days in the dark if given a carbon source such as fructose. Please see Argueta and Summers, 2005, Arch. Microbiol. 183:338-346 for more information.
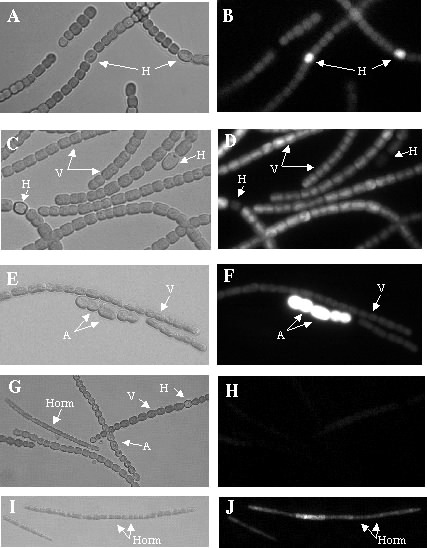
Using differential display on RNA harvested from wild-type (grows in the dark as a heterotroph) and mutant (differentiates into akinetes) allowed us to identify genes more highly expressed in akinetes after a dark shift. Promoters from these genes have been tested using the reporter plasmids shown above and have been confirmed to be more highly expressed in developing akinetes, as shown below. Please see Argueta, C., K. Yuksek, R. Patel, and M.L Summers. 2006 for more information.
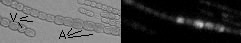
The next step in analysis is to mutate the identified gene and see if functional akinetes form in the mutant strain.
Recently initiated work: Our current research is focused on identification and characterization of akinete-related genes identified using a whole genome DNA microarray made at the UC Davis. In this work RNA from the model system as described above was hybridized to the DNA microarray. Genes that were on or off in the mutant after 3 days incubation in the dark (as compared to the wild-type strain under similar conditions where no akinete differentiation occured) were identified. Many students are now involved in confirmation of cell-type gene expression and in the construction of mutants in the identified genes. Students interested in cellular development and molecular genetics are usually involved in this type of lab project.